The Endocrine System
Communications of Yore
It's the night of the big game, and you’re the home team’s star quarterback. The whole town is in the stands. All your friends are there, plus all of their friends. You feel your stomach twist into knots as the announcer comes over the loud speaker, reading off the starting lineup.Not a sports fan? Try this one. You fret over the new pimple that emerged on your forehead this morning. You worry that a recent growth spurt has left you looking goofy, as the cuff of your shirt is now three inches above your wrist. Whether it’s pregame jitters or acne, it's not your fault. For now, blame your endocrine system for these unfortunate series of events, but be sure to make friends later, since the system controls just about every process imaginable. It's good to have on your side.
Within the endocrine system, the body is busy chatting with itself, but this time it's all about special chemicals called hormones. The endocrine system is responsible for regulating every physiological process in our bodies, from body temperature to reproduction. If you're body's doing it, odds are a hormone had something to say about it. In fact, most processes require activity from multiple hormones, multiple organs, or even crosstalk between the endocrine and nervous systems. This mode of conversation is quite complex and, well, confusing.
The main thing that defines endocrine glands is that they are ductless, meaning that hormones don’t get ejected through a duct to their target; they have direct access to the bloodstream, where they can travel long distances to perform their magic. There are also exocrine glands that work in a similar way, but they use ducts to excrete substances directly where they're needed. Gastric glands, salivary glands, and sweat glands are examples and their products, like stomach acid, saliva, and sweat, leave the gland via a duct right to the place they're needed. Exocrine glands are NOT a part of the endocrine system.
The Back and Forth of Hormones
One quirk of hormones is that some of them actually have a nemesis, an antagonistic partner that plays the exact opposite role. One hormone shows up at an organ and says, "Do this thing!" Then the other one shows up and says, "Knock it off!"For example, the pancreas is an endocrine gland that releases insulin and glucagon. Both hormones act on the liver, but they have exact opposite functions. Together they regulate blood sugar levels.
If we are stranded in the airport with nothing to eat, glucagon responds to our low blood sugar and convinces the liver to increase our blood sugar levels. If we're lucky enough to find a few Twix bars in a coat pocket, we might eat them all in a hurry. Besides having a stomachache, our blood sugar levels skyrocket and the pancreas releases the hormone insulin. Insulin tells the liver to store more sugar and lower the blood sugar levels, effectively blocking any effects that glucagon might still be having. By using the two together, and adjusting the levels as needed, the body ensures it has just the right amount of sugar in the blood, keeping things all hunky dory.
Hormones
Before we get into any specific features of the endocrine system, we've got to go over the basics: the types of hormones and the different endocrine glands. Hey, we can't put in the granite countertops or remote-controlled toilet without getting a solid foundation down first.There are about 50 chemically distinct hormones, and each end organ has a slew of receptors, each recognizing and responding to just one type of hormone. What can we say? The receptors are picky. That's not to say that only one hormone can act at any organ. Each organ has a whole team of receptors waiting for the right hormone to come along and form a match made in heaven. Hormones fall into one of three different classes: steroid, amino acid derived, and peptide.
Steroid Hormones
Steroid hormones are fat-soluble chemicals that all begin their chubby lives as cholesterol. Chemical reactions end up changing it, so each steroid hormone differs from the others based on the types, numbers, and locations of those chemical alterations. These hormones don’t normally hang out in the body. They’re made-to-order. They’re released by endocrine glands whenever the brain decides they are needed.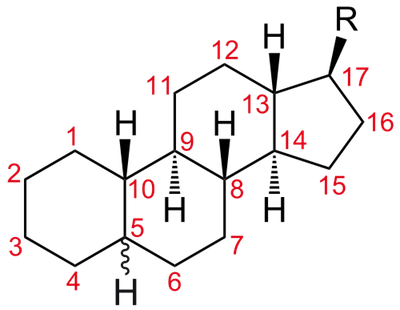
Cholesterol rings are the precursor to the steroid hormones. Each hormone will have a different chemical structure at the "R" group. Image from here.
Once the steroid hormone is synthesized and released, it travels through the blood stream to its appropriate end organ, where it's going to be put to good use addressing some particular issue. Steroid hormones are a bit of a VIP on the molecular level, and they get easy access to the inner sanctum of cells without breaking a sweat. That's because steroids are cholesterols, which are lipids. Lipids can pass right through the lipid bilayer of cell membranes, and hightail it to the nucleus where they bind to their hormone receptor. Once the steroid and receptor are linked, they travel inside the nucleus where they control the rate of protein synthesis as a team.
There are two classes of steroid hormones: corticosteroids and sex steroids. These can be further broken down into five types of steroid hormones, and each has a different and specific effect on the body.
- Corticosteriods are made in the adrenal glands and don’t regulate sexual development.
- Glucocorticoids are released from the adrenal glands and affect metabolism, decrease inflammation, and block stress.
- Mineralocorticoids are secreted from the adrenal glands and, like their name implies, help to maintain mineral levels, especially salt and water balance.
- Sex steroids are made by the adrenal glands or the gonads and regulate sexual development.
- Estrogens come from both the adrenal gland and the ovaries (the female reproductive organ that produces eggs), and promote the development and function of female sexual organs.
- Progesterone is a steroid hormone that is made in the ovaries and the placenta to regulate the menstrual cycle and maintain pregnancy.
- Androgens (made from testosterone, which we're sure you've heard of) are the male equivalent to estrogens and are responsible for developing the male sexual organs.
Amino Acid-Derived Hormones
Now on to amino acid-derived hormones. These are made up of the amino acids tyrosine, tryptophan, or glutamic acid. There are two main types of amino acid-derived hormones: thyroid hormones and the catecholamines.Thyroid hormones are released from the thyroid endocrine gland and are made up of tyrosine residues and iodine atoms. An animal can’t live without thyroid hormones, as they're involved in just about every bodily process imaginable: metabolism, growth, and development.
Catecholamines are released by the adrenal gland They're unique because they’re used by both the endocrine and nervous systems, giving them the fancy name neurohormones. These hormones regulate various processes like respiration and directing blood flow to specific organs.
Peptide Hormones
Lastly are the peptide hormones, which represent about 80% of all hormones. Peptide hormones are made from proteins. Some peptide hormones can be secreted by constitutive secretion, meaning they’re made pretty much all the time. They’re secreted from the cell as they're made and go off to do their jobs.Other peptide hormones aren’t needed around the clock. These hormones are synthesized well ahead of time and are stored in tiny vesicles until they’re needed, and then get released in a process called regulated secretion. The key difference between this and constitutive secretion is that these hormones are already baked and ready to go when needed, as opposed to being made on the spot.
Insulin is probably the best example of a peptide hormone, since it’s famous for its role in diabetes, a disorder that occurs when the body can't regulate its blood sugar levels. Insulin is made in the pancreas and stored within vesicles until you eat a Milky Way. The body digests the candy bar, and oodles of sugar make its way into your bloodstream. Then insulin is secreted to get your blood glucose levels back in check.
Both amino acid-derived and peptide hormones are water soluble, which means they can't pass through cell membranes like the lipid-soluble steroid hormones. Luckily for them, their hormone receptors aren't located inside the cells, but hang out right on the cell membrane, where the hormone can easily reach them. When the water-soluble hormone binds to its membrane-bound receptor, it triggers a signal inside the cell that affects gene transcription or protein translation. These hormones may not be able to get past the guard at the door of the cell, but they can at least make sure he passes along their message.
Endocrine Glands
There are seven different glands where hormones are synthesized and released:- Hypothalamus
- Pituitary gland
- Thyroid
- Pancreas
- Adrenal glands
- Pineal gland
- Gonads
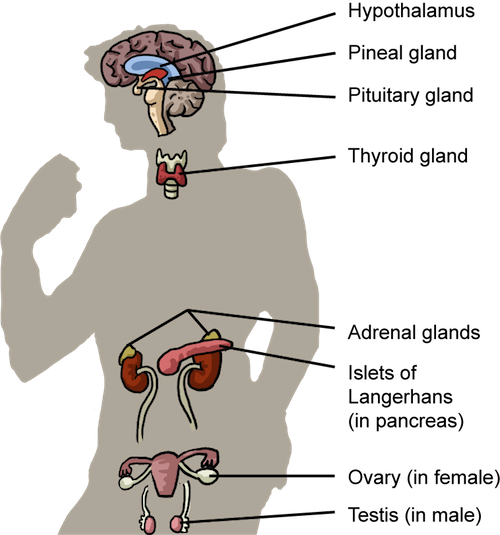
The Endocrine System. In one way or another, the endocrine system contributes to all physiological processes.
Each of these endocrine glands responds to specific stimuli and releases particular hormones. Some glands respond to hormones, others respond to a signal from the brain, and others wait for some specific change in the body before they jump into action.
The King and Queen of the Endocrine System
We'll start with the king and queen of endocrine glands: the hypothalamus and the pituitary gland. Both glands are located in the brain. Although the pituitary gland controls a boatload of hormones as the one true Glandular King, it's Her Majesty, Queen Hypothalamus, who’s really in charge. The hypothalamus tells the pituitary gland what to do and when to do it. The hypothalamus releases either stimulating or inhibiting hormones that act at the pituitary and regulate the release of its hormones.Growth hormone (the actual, for-real name of a hormone) is secreted from the pituitary gland, and controls growth and development, among other processes. The pituitary gland can't decide by itself that it wants to secrete this hormone, and so it needs a permission slip from the hypothalamus. When growth hormone is needed, the hypothalamus secretes growth hormone-releasing hormone (don't ask us where they come up with these catchy names), which activates the pituitary to release the actual growth hormone.
The pituitary gland also releases prolactin (it stimulates milk production in mammals—notice the word "lactate" kind of hiding in there), and sex hormones, including follicle-stimulating hormone (FSH) and luteinizing hormone (LH). If you're a girl, these hormones regulate ovulation. Guys have them, too, and FSH and LH help them make sperm and testosterone.
Thyroid Glands
The hypothalamus (by secreting thyroid-releasing hormone) and pituitary gland (with thyroid-stimulating hormone) also control the actions of the thyroid glands. These glands are located on both sides of the trachea (that's your throat). The thyroid glands release, you guessed it, thyroid hormone.Thyroid hormone is actually a pair of hormones: triiodothyronine (thankfully it's abbreviated T3, since it's made from the amino acid tyrosine and has 3 iodine atoms) and thyroxine (T4, since it has 4 iodine atoms). These are super important hormones, because they regulate all sorts of processes, like muscle tone, digestion, and reproduction. These hormones are also responsible for making tadpoles lose their tadpole tail to become an adult frog. Multitalented, these guys.
The Pancreas
The pancreas is next on our list of endocrine glands. The pancreas is unique, in that it has both endocrine and exocrine functions. When it has its exocrine hat on, the pancreas excretes chemicals through ducts that aid in digestion. Enzymes like lipase (to break down fats) and amylase (to break down sugars) get released when the pancreas senses we’ve just devoured a funnel cake.When the pancreas is wearing its endocrine hat, we're looking at a small region of pancreatic cells, called the islets of Langerhans. That sounds like some exotic island in the Caribbean, but don't let visions of sandy beaches carry you away. These tropical cells release both insulin and glucagon, which tag team to regulate blood sugar levels.
Adrenal Glands
The adrenal cortex and the adrenal medulla (which collectively make up the adrenal glands) are endocrine glands located on the kidney. You know that feeling you get when you lean back a bit too far in your chair, and almost flip over? Right after you save yourself, you might notice that you're breathing more heavily, and maybe even shaking and sweating a bit. You can thank this adrenal duo.Both glands release hormones that deal with stress, but they do so with differing stimuli and by releasing different types of hormones. Nevertheless, whether it's a deer's stress from running away from a not-so-effectively camouflaged hunter or a human's stress when thinking about an upcoming exam, the adrenal glands cover all things stressful, large and small.

The adrenal glands rest right on top of the kidneys. Like the brain and the kidney, the adrenal glands have two main parts: an inner medulla and an outer cortex.
The adrenal medulla responds directly to the deer's stress when it hears the hunter and releases neurohormones, like adrenaline. These are amino acid-derived hormones, made from tyrosine. By acting at adrenergic receptors (notice the "adren" in adrenal, adrenaline, and adrenergic) at various parts in the body, these hormones activate a "fight or flight" response, which is part of the nervous system. They dilate the deer's bronchioles in the lungs so it can suck down more oxygen when it's running for its life. Its heart rate increases so its muscles receive more blood (carrying helpful bits of energy) and it can run even faster. We'll learn more about neurohormones in our later section on the nervous system, so stay tuned.
The two corticosteroids, glucocorticoids and mineralocorticoids, are synthesized and released from the adrenal cortex. These hormones respond to stress in a behind-the-scenes kind of way, rather than being released in direct response to stressful situations. Hormones from the pituitary and hypothalamus stimulate the adrenal cortex.
Glucocorticoids primarily affect glucose metabolism and promote the synthesis or release of glucose into the blood. It's this added sugar that gives the deer his energy to run longer and faster than the hunter. Mineralocorticoids affect mineral metabolism, and help maintain the salt and water balance. Long story short, these corticosteroid hormones get extra energy to the muscles so the deer can outrun his predator.
Pineal Gland
The pineal gland is the next stop in our tour of endocrine glands. It's teeny tiny and just above the roof of your mouth. Melatonin is the name of the hormonal game here, and it's secreted in the absence of light. At night, and during seasons where the days are short (like winter), melatonin is released and controls the circadian rhythm, a set of biological processes that happen every 24 hours. In animals that have breeding seasons like sheep, melatonin blocks the release of certain sex hormones. In humans, it just makes us sleepy.Sex Steroids
Last, but certainly not least, we'll talk about the sex steroids released from the gonads. In men, the testes create and release androgens (including testosterone), and in women, ovaries whip up estrogen and progesterone. (Estrogen is also produced by the adrenal glands, but to a much smaller extent.)Not only do these hormones aid in sperm and egg production, but they also help develop sexual characteristics when young men and women reach that time in their lives when their voices start to crack and they become terrified of the opposite sex. These microscopic folks are responsible for turning our childhood bodies into adult male and female bodies.
The kidneys, heart, and liver are also hormone-releasing organs, but aren't usually grouped with these other endocrine glands. That's lucky for you: no test questions on them.
Hormone Cascade Pathway
Usually bodily processes don't involve just one endocrine gland and one hormone, but involve multiple glands, hormones, and end organs that talk to each other in what's known as a hormone cascade pathway. In this hormonal game of "you're it," one hormone regulates the release of another. These hormones, called trophic hormones, act on other endocrine glands, and decide whether another hormone should be released or not. Trophic hormones usually come from the hypothalamus or the pituitary gland.We'll use the prolactin hormone as an example of how hormone cascade pathways work. Prolactin is important in mammary gland development, milk production, and immune function. The pituitary gland is responsible for actual prolactin release, but other hormones control the pituitary gland's ability to secrete it. It doesn't make much sense for female mammals to be producing milk all the time. Waste not; want not (is what these organs would say if they had a mouth).
A chemical called dopamine that is secreted by the hypothalamus provides the main brake on prolactin release. It acts as a trophic hormone, binds to the pituitary gland, and blocks prolactin synthesis and release. Prolactin is a peptide hormone and has to be synthesized in advance. That means anything that messes with dopamine concentrations will also affect prolactin. Our brains are actually bathed in dopamine, so there's always dopamine around to restrict prolactin release.

Prolactin Pathway. Since female mammals don't always need prolactin, they use multiple hormones to regulate its synthesis and secretion.
If dopamine is always around, preventing the pituitary gland from releasing prolactin, how the heck is a baby calf supposed to get any milk from its mom? Well, here come a couple more hormones to make that happen.
Thyroid-releasing hormones (from the hypothalamus) and gonadotropin-releasing hormones (from the pituitary) stimulate the production and release of prolactin. They take the attention away from dopamine and put the spotlight back on prolactin. Estrogen, that steroid sex hormone, also stimulates prolactin synthesis and release.
As a mother gets late in her pregnancy, estrogen levels rise and increase prolactin levels. This increased prolactin is what prepares her mammary glands to lactate and provide her baby with milk after birth. When babies stimulate their mother's mammary glands, the hypothalamus gets involved and releases prolactin-stimulating hormones to jump-start the pituitary gland.
In the case of prolactin, there are lots of chemicals that control the synthesis and release of just one hormone from the pituitary gland. Dopamine blocks it, but other hormones stimulate it. Whichever's concentration is higher wins the battle, and ultimately decide prolactin's fate.
Brain Snack
Meet Robert Wadlow: World's Tallest Person. He had an overactive pituitary gland, and it caused excessive growth hormone secretion. When Mr. Wadlow died in 1940 at age 22 from a blister infection, he was less than an inch shy of 9 feet tall and weighed 490 pounds. He wore a size 37 shoe. That's one big dude. It was one teensy little gland stuck in overdrive that made it happen.You might be thinking that only humans are hormone driven. But don’t be fooled. All animals have endocrine systems, even ants.