Recombinant DNA
Spider-Man and Other Examples of Recombinant DNA
In the most recent Spider-Man movie, a mutant spider bit Peter Parker and passed its spider genes to him, giving him super strength, vision, and web-slinging action. Therefore, in this case, Spider-Man exists because recombinant DNA from spider genes entered the human genome of Peter Parker.If you are a Spider-Man fan, you will know the original story had Peter Parker bitten by a radioactive spider. Unfortunately, that method of transferring spider powers is highly unlikely, knowing what we know about DNA. Making recombinant DNA involves taking a gene, usually from an eukaryote, and putting it into a bacterial plasmid.
Many people fear that manipulating recombinant DNA is akin to "playing God," and some non-Spider-Man films play off those fears with the evil scientist character. Movies like Rise of the Planet of the Apes, The Island of Doctor Moreau, and Jurassic Park all feature evil scientists doing evil science with evil recombinant DNA. However, this technology has far more advantages than disadvantages, and the chances of creating super apes that will wipe out humanity are very small. Hey, we at Shmoop admit that super apes are super scary.
We will discuss the advantages and disadvantages of recombinant DNA technology in the "Biotechnology" section. For now, let's pretend we're scientists making recombinant DNA. And just for fun, let's be evil about it.
Yes! Even You Can Make a Super Ape!
So you want to make an army of super apes, but where do you start? While the following section is completely hypothetical, it shows how scientists make genetically recombinant organisms.Disclaimer: do not tell your friends after reading this that you know how to make an army of super apes. That secret will always remain in the vaults at Shmoop. We can neither confirm nor deny that Shmoop has an army of super apes.
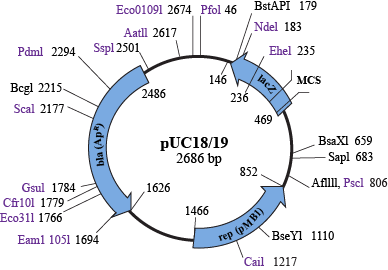
Let's make some hypothetical super apes. First, you start off with a vector, or a specific plasmid that is used for generating recombinant DNA. Vectors come in a variety of forms, but they must have three sequences:
- They need an origin of replication (oriR) site, which is a sequence that signals bacteria to replicate this plasmid whenever they replicate.
- There must be an antibiotic resistance gene, which is important for selecting bacteria that have the recombinant plasmid. When a scientist grows bacteria, he will add an antibiotic to kill all bacteria that do not have the plasmid. The antibiotic resistance gene will allow the bacteria that have the plasmid to stay alive. Score.
- A multiple cloning site (MCS) is important for facilitating insertion of the target gene into the vector. Putting the MCS into a lacZ or ccdB gene allows selection for bacteria that have the target gene inserted into the vector. This is because the target gene will disrupt the lacZ or ccdB gene, and one can easily test the activity of either of these genes by looking for colonies on a plate that lack the relevant gene activity.

A multiple cloning site is a DNA sequence that contains many restriction enzyme sites. Restriction enzymes cleave DNA at specific palindromic sequences, such as GGATCC, GAATTC, and GTTAAC. There are thousands of different restriction enzymes that each cleave a specific sequence. MCS sequences have sites for common restriction enzymes, and these enzymes were designed to cut the DNA only in the MCS and nowhere else in the vector.
Restriction enzymes cut in a way that either leaves sticky ends, where each strand is cut at a different position, leaving 2–5 bases of single-stranded DNA, or blunt ends, where the same spot on both strands is cleaved. Sticky ends are generally easier to handle, and different restriction enzymes often leave the same overhanging sequences, so you can cut your vector with one enzyme and your DNA insert with another enzyme, and the two sticky ends are then called compatible. Check out this link. Match.com could learn something from restriction enzymes.
Once you have cut your vector in the MCS, or elsewhere in the vector that has a unique restriction site, you need to join it together. How do you join the two sequences? Naturally, you join them in the same way you join everything else: glue!
Wait, not actual glue. Elmer's does not make bottles that small. Instead, we use a "molecular glue" called DNA ligase. DNA ligase is an enzyme that takes a free phosphate from the 5' end of one piece of DNA and attaches it to the free –OH at the 3' end of the other piece of DNA.
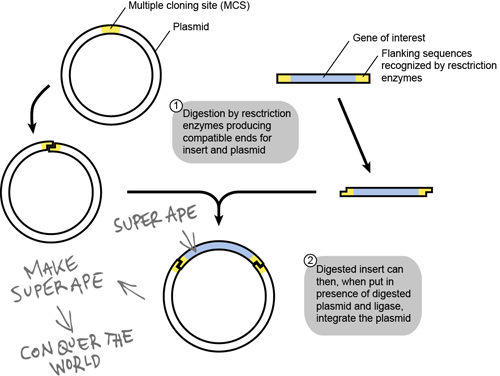
Above is a schematic of how most scientists make recombinant DNA plasmids. The target gene here is our SUPER APE gene. The only concern with making a plasmid is that there is a DNA sequence limitation. You cannot insert a DNA sequence into a traditional vector if it is over 20 kilobases (kb) in length. What if your hypothetical SUPER APE gene is huge (> 300 kb)? That will put a damper on your plot of world domination, right? Not at all, my fiendish friend!
Our scientist minions, er, friends have developed cosmids, or plasmids that can handle large pieces of DNA and contain cos sequences from viruses that infect bacteria, called bacteriophages. Cosmids can handle 50-kb inserts because they integrate into the bacterial genome rather than hanging outside of the bacterial genome, like plasmids.
If you want even more space for DNA, you can use bacterial artificial chromosomes (BACs), which handle 150–350 kb sequence inserts. BACs are similar in size to most other plasmids (5–7 kb in length), though they contain parA and parB genes, as well as oriS sites and the repE and repF genes for regulating the BAC (and its insert) so that it is properly replicated, maintained, and segregated into progeny cells. They basically act like a second bacterial genome in bacteria, and those sequences make sure your giant SUPER APE gene is replicated in the bacteria. Stick your SUPER APE gene into the BAC, and you are all set for world domination.
Brain Snack
Video on microinjection techniques
I Have Made a Plasmid: Now What?
All those months working diligently in the lab to make your SUPER APE plasmid have come to fruition (Heaven help us all…). You're ready to lead an army of super apes. But, how do you go from plasmid to super ape?There are two strategies, and it depends on the biology of your SUPER APE gene. Sorry, you need to do more research on how SUPER APE makes apes super. There are a few possibilities on why current apes are not super: either they lack the enzyme that makes them super, or they have the SUPER APE gene already but it is defective.
Let's say that apes have a dysfunctional SUPER APE gene. We can put a functional SUPER APE gene into ape embryos, and they will make the super ape gene. How does one insert a gene into an embryo? One way is to apply an electric current to the embryo, which opens the cells to take in our SUPER APE BAC. Alternatively, you can take a tiny syringe and inject the DNA into the embryo. However, both methods have several problems and are incredibly difficult to do.
There is potentially a major biological concern if we add functional SUPER APE genes to an embryo. Inserting DNA into an embryo will lead to every cell in the body having that DNA. Perhaps not every part of the body needs a functional SUPER APE gene to make the ape super. What if SUPER APE kills apes when expressed in the heart or lungs? This might be whythe gene is nonfunctional in all apes.
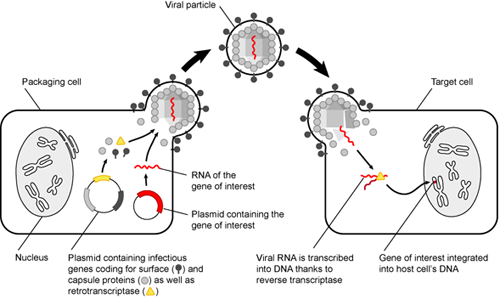
You tried adding your plasmid to embryos, and all your apes died or were never born. That is a bummer, and not just because you have a lot of dead apes on your hands now. To avoid this, we could target SUPER APE to specific tissues, such as muscles and blood cells.
There are many tissue-specific markers, or transmembrane proteins, that are expressed in certain cell types. Therefore, we can use viruses that infect cells expressing muscle and blood markers to put our SUPER APE gene in muscle and blood cells. As a disclaimer, this would only be possible if SUPER APE were a small gene (< 10 kb) using current technology. But let's assume that you are way smarter than every scientist in the world and move on. Super apes here we come!
Oops! When we put the SUPER APE gene into blood and muscle cells, it integrates into the genome and is immediately silenced by DNA methylation and tight histone packing. We need to deliver the SUPER APE gene as a protein into blood and muscle cells. Therefore, we need to make SUPER APE protein in bacteria and deliver it to our apes; however, we cannot use the same plasmid that we were using before.
"Why not?" you ask. We were using the SUPER APE gene, which includes introns, and if we are making protein, we do not want intron sequences, because bacteria do not know how to splice out introns. You will have a nonfunctional SUPER APE protein.
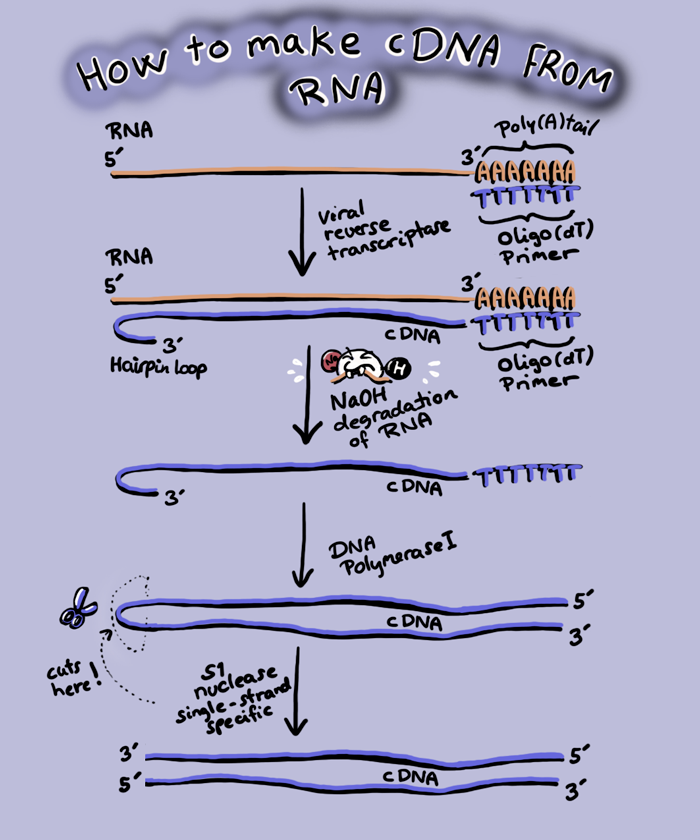
Instead of the SUPER APE gene, we want to put the cDNA (complementary DNA) of SUPER APE into a plasmid, which will be much smaller than the gene. A cDNA is a DNA made from an RNA using reverse transcriptase from retroviruses.
While every other polymerase you have heard of either makes DNA from a DNA template (DNA polymerase) or RNA from DNA (RNA polymerase), retroviruses can make DNA from an RNA template, though there are viruses that make RNA from RNA templates. Reverse transcriptase is the enzyme that makes DNA from RNA, and we can purify SUPER APE mRNA and add reverse transcriptase to make cDNA.
This cDNA is inserted into a plasmid that has a promoter to drive expression in bacteria. We can then make lots of super ape protein in bacteria. How we get it into muscle and blood cells is a trickier problem, which we will leave to you to solve (and is probably another reason why we do not already have super apes).
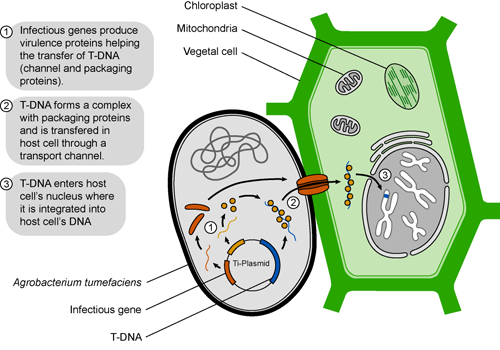
One possible way to deliver super ape proteins to blood and muscle cells would be through ingesting food that makes super ape protein. Therefore, we could have plants make super ape protein and feed those plants to our apes.
The most trusted way of delivering genes to plants for biotechnology is using Agrobacterium tumerfaciens. This bacteria infects plants and causes tumors, collectively known as crown-gall disease. Agrobacterium transfers a portion of its tumor inducing (Ti) plasmid DNA, called T-DNA,which causes the plant to make opines, or compounds that the bacterium uses for energy and carbon sources.
Scientists have used this genetic transfer as a tool for gene delivery into plants, much in the same way the super ape protein can be made in bacteria. Putting the SUPER APE cDNA into the Ti plasmid could allow plants to make the super ape protein. Our apes can eat the super-ape-expressing plants, making them super strong for our invincible army. The bonus is that if they get out of control, we stop feeding them super ape protein plants. Hopefully, they do not learn how to grow their own super ape plants. Then, it would be curtains for us all.
Can We Stop Talking About Super Apes?
Fine. We see how it is.Much of what we previously described are techniques that real scientists use to actually make proteins and deliver genes for numerous biotechnology applications. What are some real, non-super-ape applications of recombinant DNA technology?
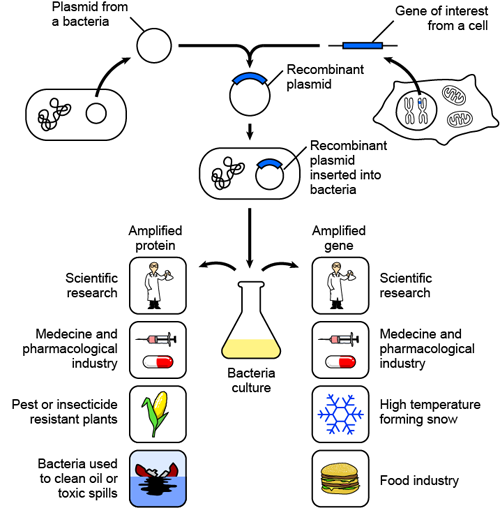
There are so many applications for recombinant DNA technology: medical applications, including gene therapy and vaccines; the creation of genetically modified crops that are resistant to pesticides or that make extra vitamins and minerals; and bacteria that can clean oil spills and even make "fake snow." While the applications of recombinant DNA technology are numerous, its limitations are its potential effects on our ecosystem. A longer discussion of this can be found in the "Biotechnology" section.
Brain Snack
Scientists use protein-making bacteria that improve the ability of water to form ice crystals. Here is a video of how scientists found those bacteria.